RESEARCH
Mechanisms and regulation of protein machines at the single-molecule and atomic level
Molecular machines are proteins that convert chemical energy into conformational changes so as to carry out work in the cell. Our group studies how the structural architectures and dynamics of molecular machines together enable their function and regulation. One major focus are nucleic-acid interacting enzymes whose aberrant function or dysregulation is often associated with severe disease states such as cancer.
In order to understand the function and regulation of these enzymes at a molecular level, we develop and apply single-molecule fluorescence methodologies and combine them with a range of structural approaches, biochemistry and computer simulations.
In order to understand the function and regulation of these enzymes at a molecular level, we develop and apply single-molecule fluorescence methodologies and combine them with a range of structural approaches, biochemistry and computer simulations.
Single-molecule biology
Knowledge of the static architecture of molecular machines alone may not satisfactorily explain how they work. Molecular machines are dynamic in nature: conformational variability (time-dependent structural fluctuations) is inherent to their mechanisms and functions.
In order to investigate this dynamic nature, we explore single-molecule imaging approaches to directly visualize molecular machines in real time. Their complex dynamics can be difficult to capture in classical bulk experiments since ensemble averaging can obscure the presence of multiple kinetic pathways or transient states. Investigations at the single-molecule level, however, can allow us to directly observe these processes (without the need for synchronization) and to correlate structural dynamics with function.
In order to investigate this dynamic nature, we explore single-molecule imaging approaches to directly visualize molecular machines in real time. Their complex dynamics can be difficult to capture in classical bulk experiments since ensemble averaging can obscure the presence of multiple kinetic pathways or transient states. Investigations at the single-molecule level, however, can allow us to directly observe these processes (without the need for synchronization) and to correlate structural dynamics with function.
Single-molecule FRET
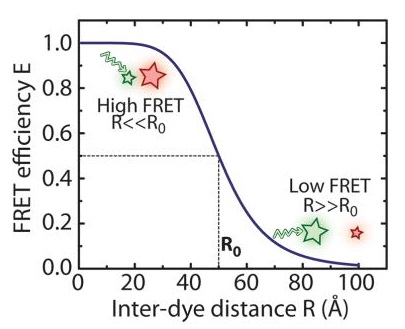
| Essential cellular processes such as interactions between biomolecules or conformational changes often involve distance changes at a nanometer length scale (1–10 nm). Fluorescence resonance energy transfer (FRET) (Forster, 1949; Stryer & Haugland, 1967) is a spectroscopic technique that enables the observation of distance changes at this length scale with high sensitivity and in real time. In this technique, a donor and an acceptor fluorophore are introduced at sites whose distance is to be monitored. Upon excitation of the donor fluorophore, a fraction of its energy can be transferred to the acceptor fluorophore in a nonradiative process. The efficiency of energy transfer, E, is highly sensitive to the distance R between the two fluorescent molecules. FRET measurements at the single-molecule level allow the observation of dynamics on a molecular scale that would be inaccessible in ensemble measurements due to random averaging (Ha, 2001; Ha et al., 1996; Zhuang et al., 2000). The ability to monitor conformational dynamics of individual molecules in real time makes single-molecule techniques highly effective in the study of a wide range of mechanistic questions. Adapted from: Deindl & Zhuang, 2012 |
|
| Essential cellular processes such as interactions between biomolecules or conformational changes often involve distance changes at a nanometer length scale (1–10 nm). Fluorescence resonance energy transfer (FRET) (Forster, 1949; Stryer & Haugland, 1967) is a spectroscopic technique that enables the observation of distance changes at this length scale with high sensitivity and in real time. In this technique, a donor and an acceptor fluorophore are introduced at sites whose distance is to be monitored. Upon excitation of the donor fluorophore, a fraction of its energy can be transferred to the acceptor fluorophore in a nonradiative process. The efficiency of energy transfer, E, is highly sensitive to the distance R between the two fluorescent molecules. FRET measurements at the single-molecule level allow the observation of dynamics on a molecular scale that would be inaccessible in ensemble measurements due to random averaging (Ha, 2001; Ha et al., 1996; Zhuang et al., 2000). The ability to monitor conformational dynamics of individual molecules in real time makes single-molecule techniques highly effective in the study of a wide range of mechanistic questions. Adapted from: Deindl & Zhuang, 2012 |
|
Integrative structural biology approach
| We study the structural architecture of molecular machines using various techniques including X-ray crystallography, cryo-electron microscopy, small angle X-ray scattering (SAXS), and cross-linking mass spectrometry. Ultimately, we hope to combine structural data and biochemistry with real-time dynamic information from single-molecule experiments in order to provide a more complete quantitative and mechanistic understanding of molecular machines. |
|











